Press Releases





Japan Agency for Marine-Earth Science and Technology
National Institute of Advanced Industrial Science and Technology
Nagaoka University of Technology
Marine Works Japan
National Institute for Natural Sciences
Capturing our closest prokaryotic relative reveals insight into the origin of Eukaryotes
1. Key points
- ◆
- Cultivation of us Eukaryotes’ closest prokaryotic (microbial) relative from deep-sea sediments
- ◆
- This prokaryote belongs to microbial lineage known as “Archaea”, possesses many genes thought to have been unique to Eukaryotes, depends on symbioses, and produces unique tentacle-like features.
- ◆
- Based on the structure and physiology, we propose a new theory (E3 model) for the evolutionary origin of Eukaryotes
2. Overview
Dr. Hiroyuki Imachi of the Japan Agency for Marine-Earth Science and Technology (JAMSTEC) and Dr. Masaru K. Nobu of the National Institute of Advanced Industrial Science and Technology (AIST), and colleagues accomplished the first cultivation and characterization of us Eukaryotes’ closest prokaryotic relative and revealed unprecedented insight into the origin of Eukaryotes.
Eukaryotes (complex organisms including animals, plants, fungi, and amoeba) are thought to have evolved from Prokaryotes (structurally simple and unicellular organisms), yet, “how” this took place remains a major mystery. Prokaryotes can be divided into two groups: Bacteria and Archaea. Current data suggests that the first Eukaryote emerged through an “archaeon” engulfing a “bacterium” and fusing into a single biological entity. The hosting archaeon is hypothesized to have belonged to a group of Archaea known as Asgard archaea. However, the appearance, way of life, and evolutionary story of these organisms remained unknown without living cells that could be studied in the laboratory.
This JAMSTEC-AIST collaboration succeeded capturing the first archaeon of this group. Cultivation was accomplished through combining the new and old – artificially recreating the deep-sea sediment environment in a unique reactor, monitoring growth using modern DNA sequencing technology, and purification through traditional cultivation techniques. A culture of the archaeon was finally reached through 12 years of strategic experimentation. The strain was named “Prometheoarchaeum syntrophicum MK-D1” after the Greek god, Prometheus, who created mankind from mud.
Though closely related to the ancestor of Eukaryotes, MK-D1 displayed many biological and physical features distinct from Eukaryotes. MK-D1 grew slowly, relied on symbiosis with other prokaryotes to grow on amino acids as energy sources, and could only grow in the absence of oxygen. Physically, MK-D1 cells were extremely small (~550 nm diameter; roughly 1/2000 of a millimeter) and simple on the inside, lacking any complex internal structures. On the other hand, MK-D1 possessed a structure never observed in prokaryotes before – branching tentacle-like protrusions extending from the cell surface.
Based on the physical, biological, and genomic characteristics unveiled by this work, the researchers propose a novel theory for the origin of eukaryotes: the “Entangle-Engulf-Endogenize (E3) model”. With the advent of photosynthesis, oxygen levels rose on Earth and created an opportunity for high-energy respiration. In the theory, the Eukaryotes’ ancestor archaeon formed symbiosis with an oxygen-respiring bacterium (the free-living ancestor of the mitochondrion), used tentacle-like protrusions and vesicles (like those observed in MK-D1) to engulf the bacterium, further developed the symbiosis internally, and matured into a single biological entity capable of digesting amino acids and breathing oxygen. This represents a primordial ancestor of us Eukaryotes.
Further research of this archaeon and its relatives will illuminate the evolutionary path structurally simple Prokaryotes took to evolve into Eukaryotes.
This study was partially supported by grants from the Japan Society for the Promotion of Science (JSPS) (KAKENHI Grants 18687006, 21687006, 24687011, 15H02419, 19H01005, 18H03367, 26710012, 18H02426, 18H05295, 16J10845, 18H04468, 18K18795), Advanced Bioimaging Support (ABiS) funded by JSPS KAKENHI Grant Number JP16H06280, and the Cooperative Study Program (19-504) of National Institute for Physiological Sciences.
The study was published in the international journal Nature on 15 January 2020 at 13:00 (EST).
*These authors contributed equally to this study.
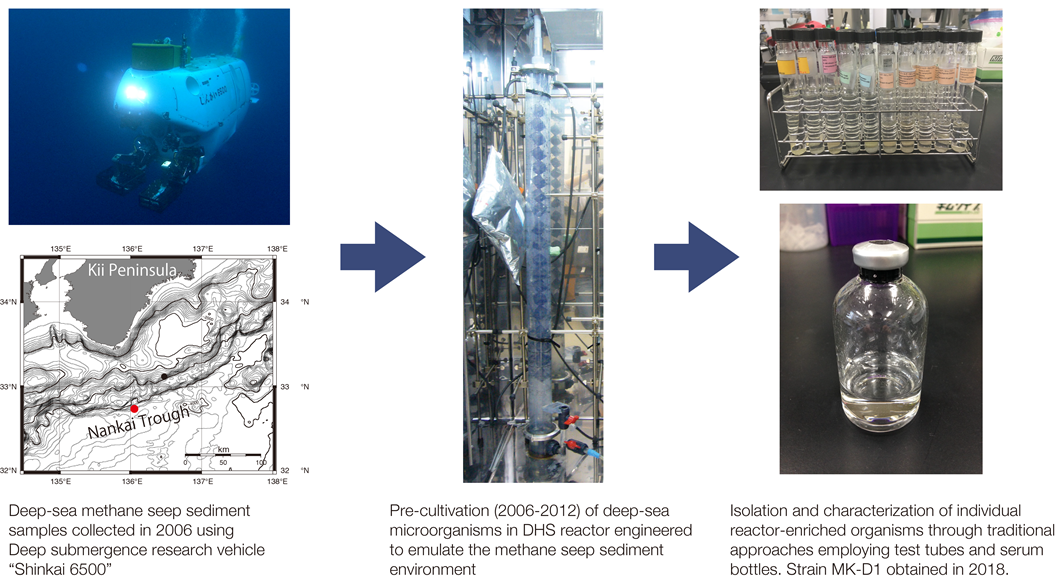
Figure 1. From marine sediments to isolation of strain MK-D1.
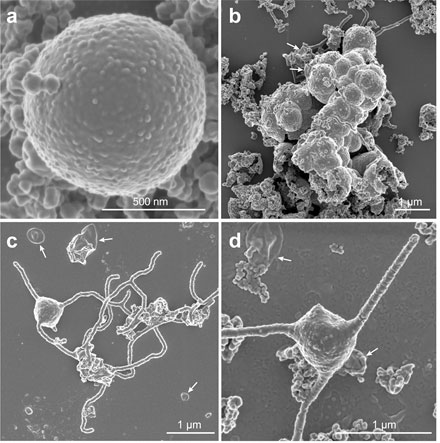
Figure 2. Scanning electron microscopy images of MK-D1 showing (a) individual cell and (b) cell aggregates in growing cultures and (c,d) cells with tentacle-like branching protrusions in cultures towards the end of growth. The cells also produce membrane vesicles (white arrows).
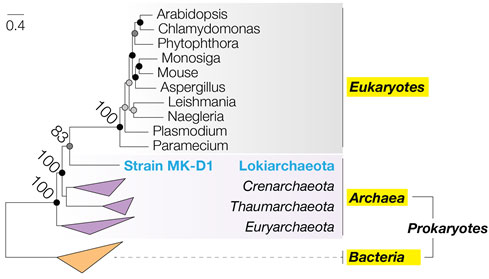
Figure 3. Phylogenetic tree showing evolutionary relationship of strain MK-D1 with Eukaryotes. The maximum likelihood tree was constructed using multiple ribosomal protein sequences found across both Prokaryotes and Eukaryotes. The values at the nodes indicate the precision (percent probability) of the evolutionary division at that node. The horizontal distance represents the evolutionary distance between individual organisms (scale at top left).
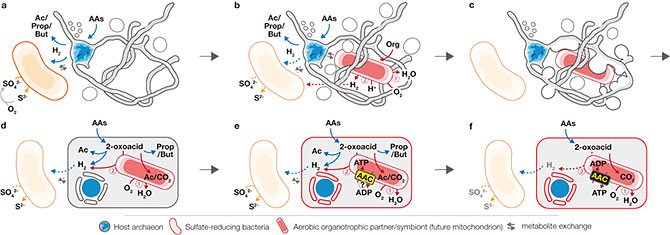
Figure 4. Proposed hypothetical model for eukaryogenesis – the “Entangle-Engulf-Endogenize (E3)” model. a, The syntrophic/fermentative host archaeon (bule cell) is suggested to degrade AAs to short-chain fatty acids and H2, possibly through interaction with H2-scavenging (and indirectly O2-scavenging) SRB (orange cell). b, The host may have further interacted with a facultatively aerobic organotrophic partner (red cell) that could scavenge toxic O2 (Continued interaction with SRB could have been beneficial but not necessarily essential; dotted arrows for interaction). c, Host external structures could have interacted (e.g., mechanical or biological fusion) with the aerobic partner to enhance physical interaction and further engulf the partner for simultaneous development of endosymbiosis and a primitive nucleoid-bounding membrane. d, After engulfment, the host and symbiont could have continued the interaction shown in b as a primitive endosymbiosis. e, Development of AAC by the endosymbiont (initial direction of ATP transport remains unclear). f, Endogenization of partner symbiosis by the host through delegation of catabolism and ATP generation to endosymbiont and establishment of symbiont-to-host ATP channel.
—the Entangle-Engulf-Endogenize (E3) model—
Contacts
- (For this study)
- Japan Agency for Marine-Earth Science and Technology
Institute for Extra-cutting-edge Science and Technology Avant-garde Research
Hiroyuki Imachi, Ph.D. (Senior Scientist) - National Institute of Advanced Industrial Science and Technology
Bioproduction Research Institute
Masaru K. Nobu, Ph.D. (Researcher) - (For press release)
- Japan Agency for Marine-Earth Science and Technology
Marine Science and Technology Strategy Department Public Relations Section - Nagaoka University of Technology
Section of General Affairs University Planning and Public Relations Office - National Institute for Natural Sciences
Research Enhancement Strategy Office