Press Releases

JAMSTEC
The Scaly-foot Snail genome sheds light on the origin of biomineralised armour
1. Key points
- ◆
- We successfully assembled the whole genome of the enigmatic Scaly-foot Snail, a deep-sea hot vent snail famed for its unique, often iron-infused, scale armour
- ◆
- We identified a set of 25 proteins (transcription factors) that regulate the making of hard parts in this enigmatic snail
- ◆
- Comparisons with data from other related animals indicate that the modification of this ancient ‘biomineralisation toolkit’ underlies the ability for animals to repeatedly invent new types of hard parts and thus obtain novel functions
2. Overview
Modern animals produce a wide range of biomineralised hard parts and skeletons, for example bones and teeth in humans, shells of snails, scales of snakes, and chaetae of various worms. These have drastically different functions ranging from structural support to feeding to protection. As such, the evolution of novel hard parts is directly linked to the diversity and acquisition of new functions, and when biomineralised elements first appeared in the Cambrian Period they led to a burst of diversification in animal body forms. Lophotrochozoan*1 invertebrates, which includes spineless animals such as molluscs (e.g. snails, clams, chitons, squids), polychaete worms, brachiopods, and bryozoans, are particularly adept at ‘inventing’ new hard structures. However, the genomic basis for these innovations have remained unresolved, although a universal ‘biomineralisation toolkit’ has been hypothesised.
The Scaly-foot Snail (Chrysomallon squamiferum; Figure 1) is unique among living snails in possessing dense dermal scales on its foot. Discovered 2001 and endemic to deep-sea hydrothermal vents in the Indian Ocean, its scales are often biomineralised with iron sulfide, making it the only known animal to use a significant amount of iron to construct its skeleton. Initially, the scales were considered to have a protective function, but recent studies indicate that they actually serve as sites for the snail to discard metabolites from its sulfur-oxidising endosymbiotic bacteria. It represents a rare case where an extensive scleritome is of a certainly recent origin (post-Jurassic), and therefore offers an ideal opportunity to test the toolkit hypothesis.
A research team led by Dr. Jin SUN (Hong Kong University of Science and Technology), Dr. Chong CHEN (SUGAR Program, X-STAR, JAMSTEC), and Dr. Norio MIYAMOTO (SUGAR Program, X-STAR, JAMSTEC) successfully sequenced the whole genome of the Scaly-foot Snail. The 444.4 Mb genome assembly is resolved at chromosomal level (Figure 1), and among the most complete and continuous among assembled mollusc or even lophotrochozoan genomes available to date (Figure 2).
By comparing genes highly expressed in tissues secreting the shell and the scales, 25 transcription factors*2 (regulatory proteins) appeared to be involved in regulating the making of hard parts in this enigmatic snail (Figure 3). Totally different subsets of these transcription factors were responsible for regulating the formation of shell and scales. Mapping these elements back to the Scaly-foot Snail genome revealed that most of them are localised on a single chromosome (‘Chr 11’ in the paper). Comparisons with an array of other lophotrochozoan animals indicated that various combinations of these transcription factors are always involved in the secretory control of all kinds of hard parts across different phyla. This suggests that these 25 transcription factors comprise the ‘biomineralisation toolkit’, which predates the origin of these lineages in the ‘Cambrian Explosion’.
These findings demonstrate that the key to inventing new hard structures is not the evolution of new genes, but instead modification and redeployment of an ancient, well-preserved suite of genomic tools. Future studies linking elements and properties of this toolkit to resulting phenotypes will shed light on how downstream processes are controlled to shape the wide variety of known hard parts, and even how to engineer new ones.
The results of this study were published in Nature Communications on April Xth, 2020 (JST). This research was supported by the Research Grants Council of Hong Kong (No. 16101219) and a Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (18K06401).
*These authors contributed equally to this work.
- *1:
- Animals belonging to the clade Lophotrochozoa, including a wide range of phyla such as Mollusca (snails, octopus, chiton, clams, slugs, etc.), Annelida (earthworms, polychaete worms, etc.), Nemertea (ribbon worms), Brachiopoda (brachiopods), among others.
- *2:
- Proteins that control/regulate the transcription of information from DNA to messenger RNA. They are able to turn genes on or off and the levels of transcription to ensure correct amount of expression takes place for a specific genes in a specific cell at the right time.
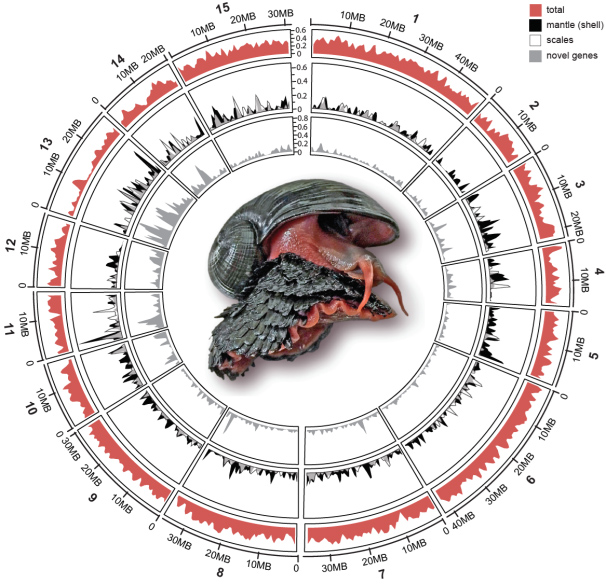
Figure 1: Key features of the Scaly-foot Snail genome. Circos plot showing the 15 chromosomal-level linkage groups. The outer ring (red peaks) indicates gene density in each pseudo-chromosome, and the inner rings shows the normalized density of the highly expressed genes.
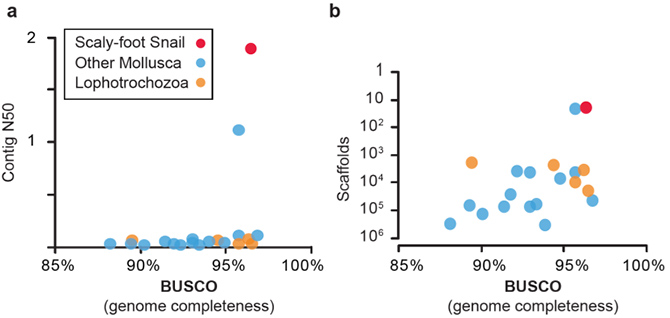
Figure 2: Quality comparisons between the Scaly-foot Snail genome and other available lophotrochozoan genomes: a, Contig N50 vs BUSCO; b, Number of scaffolds vs BUSCO
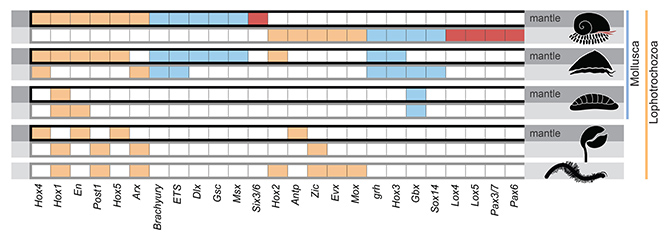
Figure 3. The ‘biomineralisation toolkit’ – transcription factors shown to be involved in armature secretion in the Scaly-foot Snail, compared to other major lophotrochozoan animals including conchiferan molluscs (bivalves, gastropods, cephalopods), aculiferan molluscs (chitons), brachiopod, and annelids. The top row for each group (darker shading) shows records of significant expression in shell-secreting mantle and bottom row (lighter shading) shows expression for other hard parts such as scales.
Contacts
- (For this study)
- Chong CHEN, Researcher, X-STAR, JAMSTEC
Norio MIYAMOTO, Researcher, X-STAR, JAMSTEC - (For press release)
- Public Relations Section, Marine Science and Technology Strategy Department, JAMSTEC