Press Releases
March 17, 2015
JAMSTEC
Successfully Inhibit Cell Proliferation
by Exploiting Vulnerability of Gene Expression System
~Potential for Application in Medical Fields~
Overview
Dr. Hideki Kobayashi, Research and Development Center for Marine Biosciences, the Japan Agency for Marine-Earth Science and Technology (JAMSTEC: Asahiko Taira, President) artificially synthesized and expressed green fluorescent protein (GFP)*1 with low-usage codon*2 in bacteria, cancer cells and viruses (protein synthesis based on genetic information). As a result, it succeeded in inhibited proliferation of those cells non-specifically.
Accurate DNA replication is essential for normal cell proliferation. In the process of gene expression, the genetic information stored in DNA is transcribed into messenger RNA (mRNA) and then translated into protein. Codons are required to specify the sequence of amino acids to make proteins. There are various types of codons, however, the codon usage is biased. Low-usage codons have a genome-wide distribution at a very low frequency. Their transfer RNAs (tRNAs), which transport amino acids corresponding to codons, occur in the cell at lower concentrations than those for normal codons. Thus, it is known that molecular interactions (interactions of biopolymers such as proteins composing human body) are very thin between low-usage codons and tRNAs with low concentration compared to those for normal codons. This can be considered as vulnerability (or may be referred to as “Achilles heel”) in gene expression system.
Focusing on this vulnerability, Dr. Kobayashi constructed an artificial gene containing many low-usage codons to exploit this security hole by monopolizing multiple minor codon tRNAs through its expression. It resulted in suspension of almost all translation in the cell, which successfully provided a novel nonspecific virus defense system in human cells.
These results, though it is still at an initial research stage at the cellular level, indicated that low-codon usage could be used for modifying non-bioactive genes to have nonspecific antitumor or antiviral activity. By carrying out further verification in cooperation with medical research institutions, it has a potential for application in gene therapy for treating rare diseases, virus infections, and cancers in the future.
These study results have been posted on a scientific journal, Applied and Environmental Microbiology on March 13, 2015 (JST).
Title: Inducible suppression of global translation by overuse of rare codons
Author:Hideki Kobayashi1
1. Research and Development Center for Marine Biosciences, JAMSTEC
A patent was obtained for this study result.
Patent No5396071: How to inhibit cell proliferation
Patent Owner: JAMSTEC
*1 Green fluorescent protein (GFP): The green fluorescent protein (GFP) is a protein that exhibits bright green fluorescence. In molecular biology, it is often used as a reporter gene.
*2 Low-usage codon: It is a type of codons which is used less frequently. A codon is a sequence of three successive bases of the mRNA, which includes adenine (A), cytosine (C), guanine (G) and uracil (U). Each codon encodes for a specific amino acid, and then the information is carried for protein synthesis. Codons have corresponding amino acid types, however, multiple codons can be encoded for the same amino acid. The codon usage is, therefore, varied and biased depending on organisms.
*3 Vulnerability: Defects in systems. It refers to a weakness in computer systems and programs attacked by computer viruses and hackers.
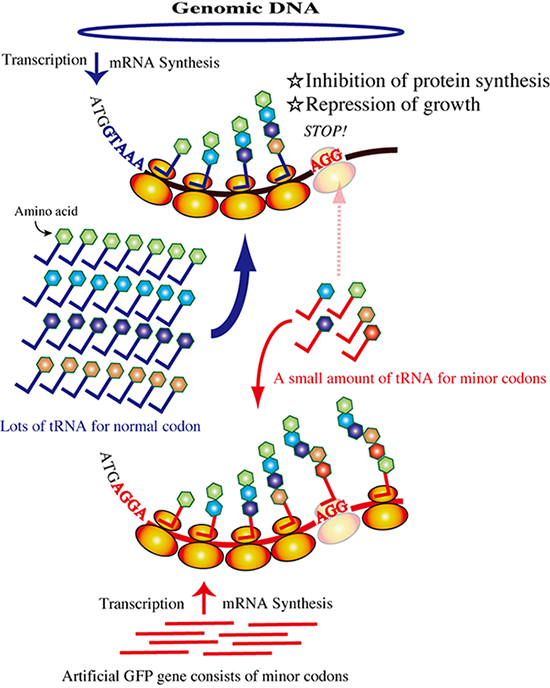
Figure 1: Schematic of posttranscriptional inhibition by the rare-codon gene.
Rarecodon tRNAs are monopolized by overexpression of the artificial gfp gene.
Translation of the mRNA transcribed from the genome is inhibited because of the shortage of rare-codon tRNAs. All cellular protein synthesis except that of GFP will be inhibited, and cellular activities such as cell growth will be reduced.
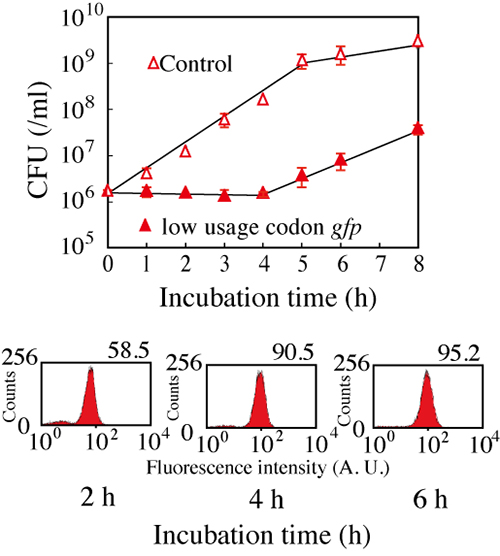
Figure 2: Suppression of Escherichia coli growth with the expression of gfp gene.
The expression of gfp gene suppressed the growth of Escherichia coli for about four
hours (the above). On the other hand, GFP expression was indicated with observation
of fluorescence (the below).
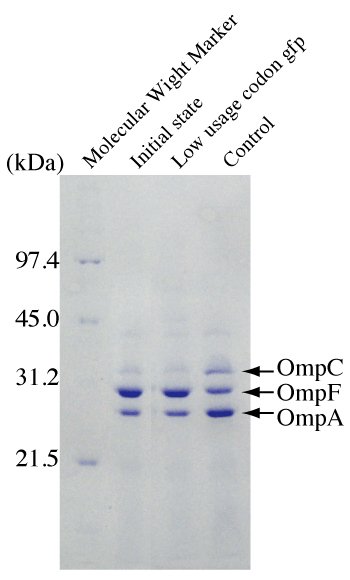
Figure 3: Effects of induction of lgfp on the expression of Escherichia coli OmpCand OmpF.
Outer membrane proteins were prepared from cell samples. Escherichia coli carrying pLGFP was grown in modified LB not containing NaCl (lane 1), and cells were harvested and incubated in LB medium (1% NaCl) with 2mMIPTG (lane 2) or without IPTG (lane 3) for two hours.

Figure 4: Effects of lgfp expression on the rates of expansion of various phages.
Escherichia coli JM2.300 carrying pHGFP or pLGFP was used as a host for phageλ, and Escherichia coli AK4 carrying pHGFP or pLGFP was used as a host for phages f1 and MS2. Phages were added to the cultures at a multiplicity of infection of 0.01. The rate of expansion of each phage was calculated from the phage titer after 4hours of incubation at 37°C. Error bars show standard deviations (n=3).
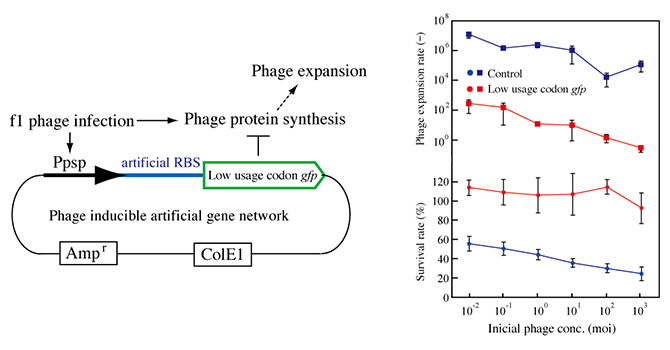
Figure 5: Construction of a phage response genetic system.
pPAL contains the lgfp gene downstream of the modified promoter of the psp operon, the ampicillin resistance gene, and ColE1. pPAH has hgfp instead of lgfp of pPAL. RBS, ribosome binding site (left).
Phage resistance of E. coli AK4 carrying pPAL. Cultures of Escherichia coli AK4 carrying pPAH (blue) or pPAL (red) were incubated with various phage concentrations at 37°C for 4 hours. Growth is indicated as the ratio of the OD to that of a culture of AK4 without the addition of phage f1 (circles, lower graph). All cultures without phage addition were grown to anOD of 0.65 to 0.90. The titers of phage f1 in the cultures were measured. The reproductive rates were calculated from the titers (squares, upper graph). Error bars show standard deviations (n=3).
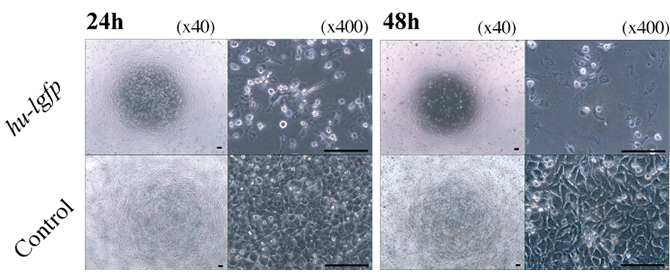
Figure 6: Effect of hu-lgfp induction on the growth of human cells and adenovirus infection.
Microscopic observation of the growth of HeLa cells transfected with pTRE-Luc or pTRE-G1. HeLa cells transfected with pTRE-Luc were used as a control.
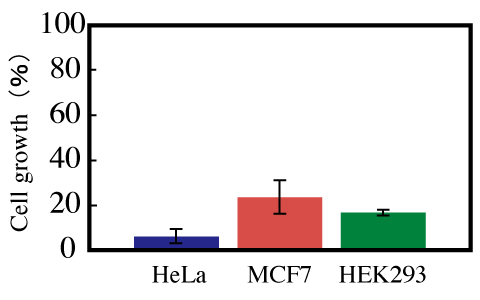
Figure 7: Effect of pTRE-G1 on the growth of HeLa, MCF7, and HEK293 cells. All cells were incubated for 48 hours. The ratio of the growth of cells transfected with pTRE-G1 to the growth of cells transfected with TRE-Luc was calculated.
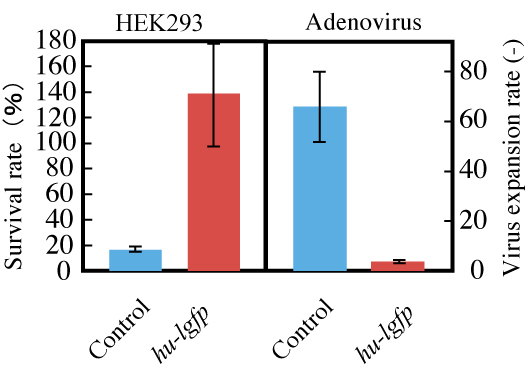
Figure 8: Effect of pTRE-G1 on recombinant adenovirus infection.
HEK293 cells grown in DMEM for 48 hours after transfection were used as hosts. Recombinant adenovirus was added to each culture well.
Cells and recombinant adenovirus particles were counted after 72 hours of cultivation.
Contacts:
- (For this study)
-
Hideki Kobayashi, Senior Scientist,
Research and Development Center for Marine Biosciences - (For press release)
- Kazushige Kikuchi, Manager, Press Division, Public Relations Department