Press Releases

JAMSTEC
University of Tsukuba
Discovery of a previously unknown virosphere hidden
inside marine microorganisms
1.Overview
A large variety of RNA viruses infect and coexist with marine microorganisms, according to a study using a novel technique for comprehensive detection of RNA viruses, called fragmented and primer ligated dsRNA sequencing (FLDS).
Viruses generally infect a host organism, where they replicate, and then, burst out, destroying the host cell to find a new host organism (Figure 1). To investigate the diversity and ecology of viruses in environments, scientists usually analyze virus particles that have burst out of cells. Viruses are primarily classified as DNA viruses, RNA viruses, and retroviruses. It has been shown that a large variety of DNA viruses exist in marine environments (Figure 2). In contrast, the diversity and ecology of RNA viruses in the natural environment are yet to be known because there is no established method of environmental genomics.
To evaluate the diversity of RNA viruses in the marine environment, our research group led by Dr. Syun-ichi Urayama, Postdoctoral Researcher (currently Assistant Professor at Department of Life and Environmental Sciences, University of Tsukuba) and Dr. Takuro Nunoura, Director-General at the Research and Development Center for Marine Biosciences developed a novel technique to comprehensively analyze genomes of RNA viruses. The results showed that a total of 842 species of RNA viruses were identified in a total of 10 liters of seawater, most of which are new species. Some are classified into half of the known RNA virus families, while others are classified into potential novel families that have never been identified (Figure 3). In addition, scientists compared the diversity of RNA viruses found in microorganisms and those found floating in seawater. Many of the viruses were identified only within microbial cells. These results indicate that the viruses detected only in microorganisms coexist with the host microorganism, instead of destroying the cells and bursting out (Figure 4).
Traditionally, most viruses were considered to be pathogenic; however, recent studies have reported some cases in which organisms infected with viruses have shown no symptoms. According to these results, it is very likely that in the natural world, many viruses co-exist in harmony with their hosts, whereas the pathogenic viruses that adversely affect the hosts seem to be a unique group (Figure 5).
By allowing comprehensive detection of RNA viruses, the novel FLDS technique is expected to contribute to biological and biotechnological advances over a wide range of applications and not just confined to the ecology of viruses. A study has already begun to explore the potential application of the FLDS technique in medicinal, public health, and agricultural fields.
This work was supported by JSPS KAKENHI Grant Numbers JP26892031, JP16H06429, JP16K21723 and JP16H06437.
The above results were published in Molecular Ecology Resources on September 7, 2018 (JST).
Title: Unveiling the RNA virosphere associated with marine microorganisms
Authors: Syun-ichi Urayama1,2, Yoshihiro Takaki1,3,4, Shinro Nishi1,4, Yukari Yoshida-Takashima3, Shigeru Deguchi1, Ken Takai3, Takuro Nunoura1,4
Affiliations: 1. Research and Development Center for Marine Biosciences, JAMSTEC
2. Laboratory of Fungal Interaction and Molecular Biology (donated by IFO), Department of Life and Environmental Sciences, University of Tsukuba
3. Department of Subsurface Geobiological Analysis and Research, JAMSTEC
4. Ecosystem Observation and Evaluation Methodology Research Unit, Project Team for Development of New-generation Research Protocol for Submarine Resources, JAMSTEC
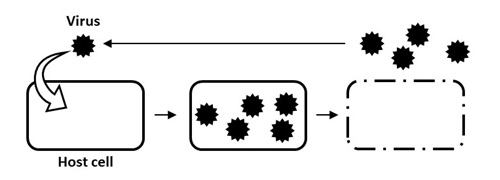
Figure 1. General life cycle of a virus
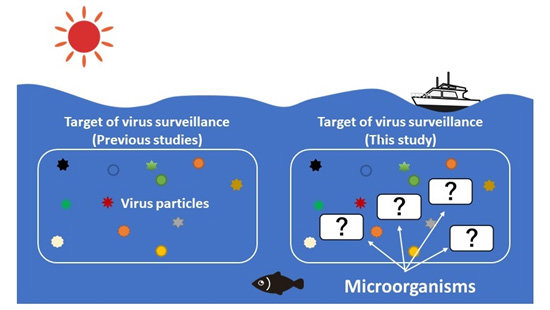
Figure 2. Traditionally detected viruses in the ocean (floating viruses) and viruses detected in this study (floating viruses + microorganisms).
Generally, virus particles floating in seawater are targets of environmental virology, which has shown that a variety of DNA viruses exist in marine environments (Left). To examine the diversity of RNA viruses, this study carried out comprehensive analysis of RNA genomes for viruses inside microorganisms and virus particles floating in the seawater (Right).
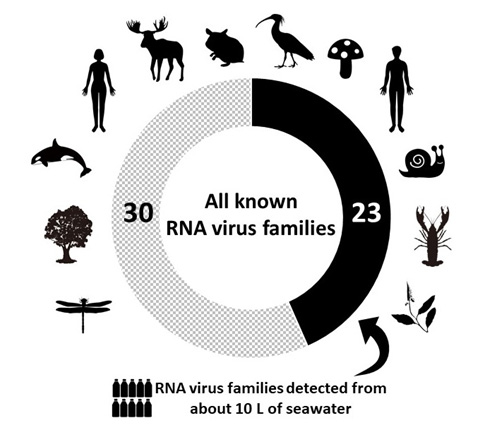
Figure 3. Half of the known RNA virus groups were found in a total of 10 liters of seawater studied.
The detected species of RNA viruses (842) were analyzed based on sequence similarity with known RNA viruses. A total of 23 known RNA virus families from among 53 RNA viral families that have been identified so far were found in this study.
In addition, several RNA viruses detected in this study belonged to completely new families, different from any of the known RNA virus families.
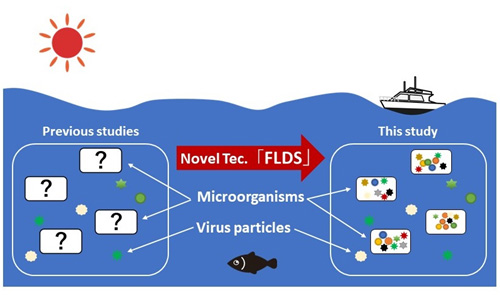
Figure 4. This study suggests that most of RNA viruses coexist in microbial cells. Only a few RNA virus groups were found in pelagic waters (Left). However, this study showed that a variety of RNA viruses do exist in the microorganisms (Right).
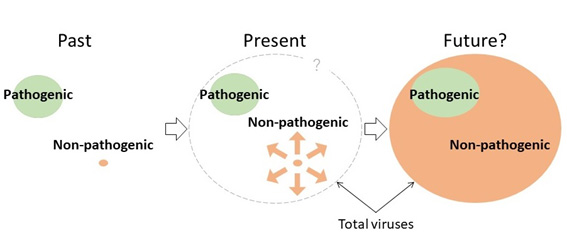
Figure 5. Do non-pathogenic viruses dominate in the virosphere?
Since the discovery of viruses as pathogens in humans, animals, and plants, most of the viruses have been identified as pathogens. However, recent studies have reported the organisms with no obvious symptoms harbored hidden viruses. This study suggests that various RNA viruses likely co-exist with their host in harmony.
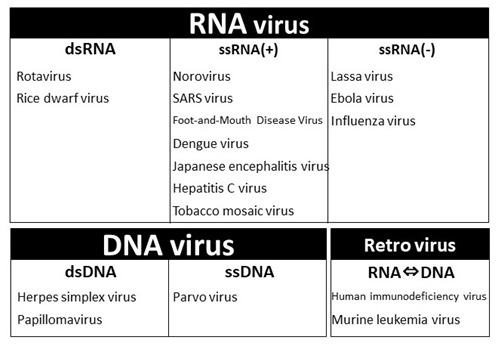
Figure 6. Classification of viruses based on genomic nucleic acid and replication method
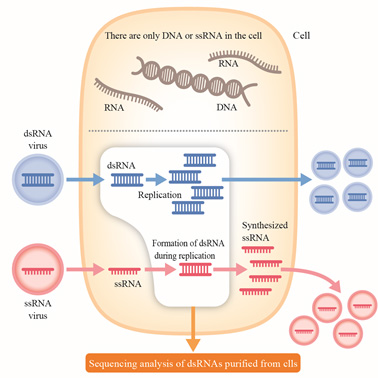
Figure 7. Principle of an efficient search system of RNA viruses hidden inside biological cells
Genomic double-stranded DNA, or single-stranded RNA such as ribosomal RNA and messenger RNA exist in healthy cells, while almost no long double-stranded RNA does. If the cell is infected with an RNA virus, long double-stranded RNA is accumulated in the cell, irrespective of the virus genome being double stranded or single stranded. The FLDS method allows effective detection of RNA viruses hidden in the cells by deciphering the sequence of long double-stranded RNA from an organism.
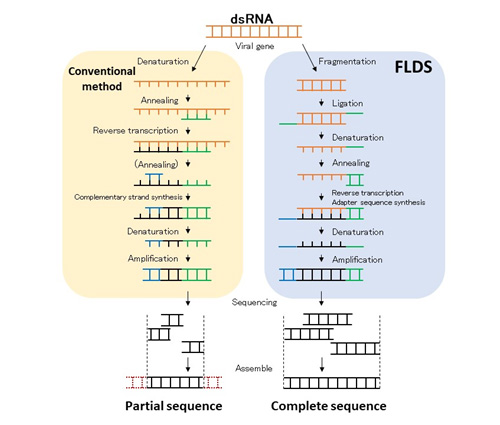
Figure 8. RNA virus information obtained by conventional methods and by FLDS
When we try to detect viruses in a comprehensive manner, it is often necessary to decipher large quantities of genetic information. Technically, this sequencing method allows us to obtain partial sequence information of the virus genome only. However, FLDS can provide full-length sequence information of double-stranded RNA from biological samples. This information is of extremely high quality.
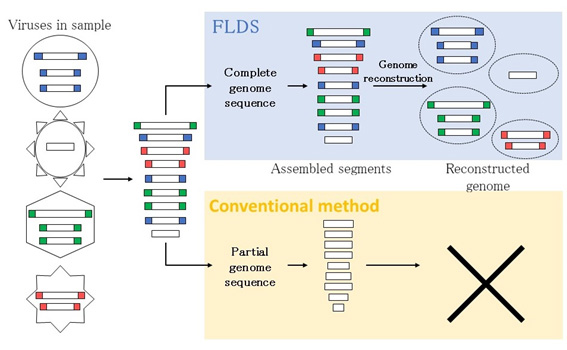
Figure 9. FLDS can reconstruct the viral genome and detect unknown virus genes.
In most cases, the genome of an RNA virus is made up of several linear nucleotide segments, two ends of which show similar sequences, as shown in colored parts of the above illustration. It is, therefore, possible to reconstruct the original complete genome set based on the full-length sequence information of the genome segment obtained by FLDS.
Contacts:
- (For this study)
- Takuro Nunoura, Deputy Director-General, Research and Development Center for Marine Biosciences
- (For press release)
- Tsuyoshi Noguchi, Manager, Press Division, Public Relations