
December 16, 2010
Japan Agency for Marine-Earth Science and Technology
Genome Sequence of Uncultivated Archaeon from Subsurface Mine
-Novel insight into the relationship between Archaea and Eukarya-
Summary
Composite genome sequence of a thermophilic archaeon (Caldiarchaeum subterraneum) that occurred in a subsurface geothermal stream and represents uncultivated archaeal lineage was reconstructed from metagenomic analysis (*1). The most striking feature of the archaea (*2) is the presence of a eukaryote-type ubiquitin-like protein modifier system that is structurally distinct from the prokaryote-type systems recently found in some prokaryotes. Ubiquitin protein modifier system is necessary for eukaryotic cellular mechanisms, and has been recognized as the hallmark of Eucarya. The presence of such a eukaryote-type system is unprecedented in prokaryotes, and indicates that a prototype of the eukaryotic protein modifier system was present in the last common ancestor of Archaea and Eucarya. The genome was also distinct to previously reported archaeal phyla/divisions, and the unique traits suggest that the uncultivated archaeal lineage represented by C. subterraneum can be considered as a novel phylum/division.
This study was carried out by Takuro Nunoura and his colleagues from the Japan Agency for Marine-Earth Science and Technology (JAMSTEC), as well as scientists from Keio University, The University of Tokyo, and Kyoto University.
The study was published online in the journal Nucleic Acids Research on December 15, 2010.
- Title:
- Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal division
- Authors:
- Takuro Nunoura1, Yoshihiro Takaki1, Jungo Kauta1, Shinro Nishi1, Junichi Sugahara2,3, Hiromi Kazama1, Gab-Joo Chee1, Masahira Hattori4, Akio Kanai2,3, Haruyuki Atomi5, Ken Takai1, Hideto Takami1
1. Extremobiosphere Research Program, Institute of Biogeosciences, Japan Agency for Marine-Earth Science & Technology (JAMSTEC), 2. Institute for Advanced Bioscience, Keio University, 3. Systems Biology Program, Graduate School of Media and Governance, Keio University, 4. Center for Omics and Bioinformatics, Graduate School of Frontier Sciences, The University of Tokyo, 5. Department of Synthetic Chemistry and Biological Chemistry, Graduate School of Engineering, Kyoto University
Backgrounds
It has been revealed that many uncultivated lineages of prokaryotes (Archaea and Bacteria) inhabit every niche on this planet. On the other hand, metagenomics has been developed after 1990s, and some genome sequences from some of uncultivated microbes have also been reported by metagenomic approach in this decade.
Candidatus ‘Caldiarchaeum subterraneum (C. subterraneum)’ represents one of the uncultivated archaeal lineages that have been detected from deep-sea hydrothermal environments, and subsurface and surface terrestrial hot-springs as ribosomal RNA gene sequences (Fig. 1). The archaeal lineage has never been cultivated by any techniques, and we did not know any of their metabolism before the metagenome project.
Ubiquitin-proteasome system (*3) that is responsible for degradation of damaged or unnecessary proteins is necessary for eukaryotic cellular mechanism, and has been observed even in primitive mono-cellular eukaryotes. Recently, prokaryote-type protein degradation systems that include prokaryote-type ubiquitin-like proteins and prokaryote-type proteasome have been discovered, but they are distinct from eukaryote-type systems, and no eukaryote-type system has been observed in any prokaryotes. Thus, eukaryote-type ubiquitin-proteasome system has still been one of the hallmarks of Eucarya.
Summary of methods
Genomic DNA directly extracted from microbial community in geothermal water stream at a subsurface mine was transformed into E. coli. We selected clones encoding putative archaeal genome fragments. Archaeal genome fragments were analyzed by a pyrosequencing technique (Fig 2).
Results and discussion
A composite genome of C. subterraneum consist of 1,680,938 bp was successfully reconstructed and a total of 1,777 genes were identified. The genome size is as much as that of previously analyzed genomes from thermophilic archaeotes, and the human genome is 1500 holds more than that of the archaeon. Topics found in this study are described as follows.
- a.
- C. subterraneum is likely a chemolithoautotrophic thermophile that utilizes hydrogen as an electron donor, and oxygen and nitrate as electron acceptors, and fixes carbon dioxide.
- b.
- C. subterraneum is distinct from previously characterized archaeal phyla/divisions in its genomic feature, and the unique traits suggest that the uncultivated archaeal lineage represented by C. subterraneum can be considered as a novel phylum/division.
- c.
- C. subterraneum has a gene cluster of eukaryote-type ubiquitin-proteasome system. The gene sequences are distinct from those of prokaryote type systems, and any signatures of lateral gene transfer from eukaryotes have not been recognized. Consequently, we concluded that eukaryote-type ubiquitin-proteasome system already occurred in the last common ancestor of Archaea and Eucarya, and most of the archaea have lost the system.
Future study
The eukaryote-type ubiquitin-proteasome system found in C. subterraneum likely preserves primitive catalytic system. We will investigate the ubiquitin-proteasome system found in C. subterraneum biochemically, and its catalytic system will be compared to that found in eukaryotes. Other unique features of C. subterraneum will also be analyzed biochemically. Thermostable enzymes from this archaeon are potential resources for biotechnological applications, including regent and food industry.
*1. Metagenomics
Metagenomics studies environmental DNA directly extracted from microbial communities without isolation using cultivation or culture-independent techniques. Usually, Polymerase chain reaction (PCR) gene amplification for specific genes from environmental DNA is not regarded as metagenomics.
*2. Archaea
One of the three domains of life; Archaea, Bacteria and Eucarya. Until recently, only two phyla, the Crenarchaeota and the Euryarchaeota, have been recognized after the establishment of the domain Archaea. Although regarded as members of the Crenarchaeota based on small subunit rRNA phylogeny, environmental genomics has recently revealed two novel phyla/divisions in the Archaea; the “Thaumarchaeota” and “Korarchaeota” (Fig. 1).
*3. Ubiquitin-proteasome system
Damaged or unnecessary proteins are modified by small ubiquitin protein, and ubiquitin is a marker for protein degradation by proteasome. Proteasome is a huge proteinase complex, and ubiquitin is released from modified proteins before protein degradation in proteasome by a deubiquitinase subunit.
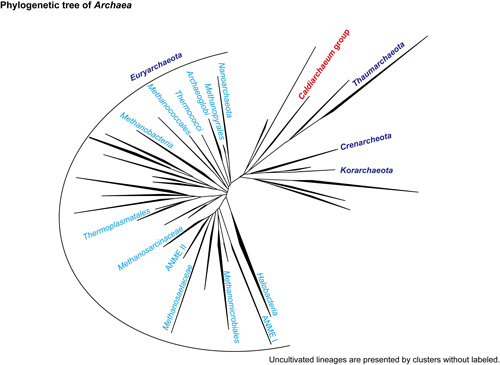
Figure 1: Uncultured lineages of archaea
Phylogenetic tree of archaea. The archaea of interest in this study belong to the group in red. Each tree branch represents phylum, class or order in the taxonomy.
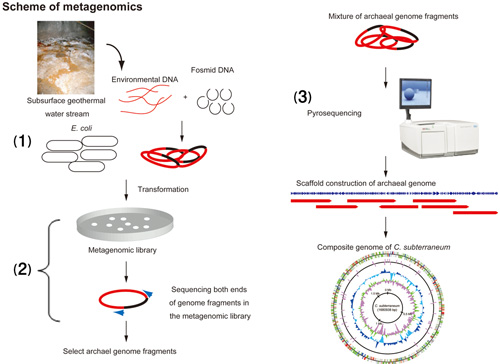
Figure 2: Scheme of metagenomics
(1) Genomic DNA directly extracted from microbial community in geothermal water stream at a subsurface mine was ligated into fosmid (mini circle sequence) and transformed into E. coli. (2) The end sequence of the genome fragments in fosmids were analyzed to select clones containing achaeal genome fragments. (3) Archaeal genome fragments were analyzed by a pyrosequencing technique.
Contacts:
- Japan Agency for Marine-Earth Science and Technology
(For the research) - Takuro Nunoura, Senior Scientist
Extremobiosphere Research Program
Institute of Biogeosciences (Biogeos)
- (For publication)
-
Toru Nakamura, e-mail: press@jamstec.go.jp
Manager, Planning Department Press Office