

January 27, 2012
JAMSTEC
Genomic Analysis of Deeply Branching Thermophile
Provides Clues to Early Life on Earth
Overview
Metagenomic analysis*1) of the microbial community in the hot stream of a Japanese epithermal mine revealed that Candidatus 'Acetothermus autotrophicum' (Ca. 'A. autotrophicum'), an uncultivated thermophilic bacterium, was keeping an ancient metabolic pathway for CO2 fixation and energy generation, known as acetyl-CoA pathway *2). The acetyl-CoA pathway is considered to have been inherited from the last universal common ancestor (LUCA) *3), a cell from which all life has evolved. Bacteria and Archaea are believed to have evolved from LUCA.
Phylogenetic trees based on proteins conserved in bacteria and archaea showed that Ca. 'A. autotrophicum' is very near to their common ancestor. Ca. 'A. autotrophicum' also retains a thermostable bifunctional fructose 1,6-bisphosphate (FBP) aldolase/phosphatase, a gluconeogenic enzyme assumed to work effectively in the early Earth environments. This suggests that Ca. 'A. Autotrophicum' is the archaeorganism still retaining functional properties of early life on Earth.
These results will considered to become direct evidence showing the existence of the functional traits in the first free-living (autotrophic) bacterial and archaeal ancestors, and provide new insights into the dawn of life to the earliest phases of evolution from the genomic point of view.
The study was published on January 27, 2012 in the online journal of Public Library of Science ONE (PLoS ONE), vol. 7, e30559 by Hideto Takami et al.
- Title:
- A deeply branching thermophilic bacterium with an ancient acetyl-CoA pathway dominates a subsurface ecosystem
- Authors:
- Hideto Takami1*, Hideki Noguchi2, Yoshihiro Takaki1, Ikuo Uchiyama3, Atsushi Toyoda4, Shinro Nishi1, Gab-Joo Chee1, Wataru Arai1, Takuro Nunoura1, Takehiko Itoh2, Masahira Hattori5, and Ken Takai1
- 1.
- Japan Agency for Marine-Earth Science and Technology (JAMSTEC)
- 2.
- Department of Biological Sciences, Tokyo Institute of Technology
- 3.
- Laboratory of Genome Informatics, National Institute for Basic Biology, National Institutes of Natural Sciences
- 4.
- National Institute of Genetics
- 5.
- The University of Tokyo
Introduction
Deep-sea hydrothermal vents are considered as a possible cradle of early life. In recent studies, specific types were considered to be the most plausible places, including low temperature alkaline H2-rich hydrothermal vents. This has led to a scenario in which the energy and cell carbon are synthesized through acetyl-CoA pathway with hydrogen and CO2 in the early Earth environments (Fig. 1). Yet biological evidence to support this hypothesis is still scarce, leaving many questions unanswered. These include processes of energy metabolism in the early branching archaeal and bacterial species.
Results
Researchers focused on microbial mat flourishing in a hot stream (70 °C) of an epithermal mine similar to the conditions in the low-temperature hydrothermal vents. The genome sequence of Ca. 'A. autotrophicum' was nearly completed and revealed the presence of the acetyl-CoA pathway, an ancient metabolic pathway believed to be possessed by the last common ancestor of Achaea and Bacteria. A Phylogenetic analysis, using a concatenated amino acid sequence of four proteins common across all prokaryotes *4), revealed that Ca. 'A. autotrophicum' is very near to LUCA among all presently known bacteria (Fig. 2).
Furthermore, Ca. 'A. autotrophicum' was found to possess fructose 1,6-bisphosphate (FBP) aldolase/phosphatase, a bifunctional enzyme indispensable for synthesizing glucose from acetyl Co-A and present only in the limited prokaryotic species thought to be very near to LUCA. Usually, enzymes catalyze both gluconeogenesis and glycolysis. FBP aldolase/phosphatise, however, catalyzes only in the direction of gluconeogenesis and is thermostable even at 70°C.
This implies that under the organic matter-poor conditions of early Earth, microbes produce acetyl-CoA from hydrogen and CO2 via the acetyle-CoA pathway, convert acetyl-CoA to acetate to acquire energy, and synthesize the saccharides by using aldolase/phosphatase for the formation of cell carbons (Fig. 4).
*1. Metagenomic analysis
Generally, it is very difficult to recover microbes from the natural environment using currently available techniques. In fact, less than 10% of the microbes can be merely cultured in the laboratory settings. Metagenomic analysis is a culture-independent method to analyze the comprehensive genomic information from microbial community to figure out the functional potential and taxonomic properties in the microbial community by using genomic DNAs directly extracted from the environmental samples without cultivation.
*2. Acetyl-CoA pathway
A metabolic pathway leading to the fixation of CO2 as acetyl-CoA, an important molecule in metabolism. It occurs under anaerobic conditions. The acetyl-CoA pathway is now found in only limited bacterial and archaeal species. Using this pathway, bacteria acquire energy by producing acetate and archaea acquire energy by producing methane.
*3. Last universal common ancestor
The last universal common ancestor (LUCA) is the most recent common ancestor of all currently living organisms believed to have appeared about 3.9 billion years ago.
*4. Prokaryote
Prokaryotes are single-celled organisms without a cell nucleus. They basically refer to bacteria and archaea. Archaea are the third domain of life discovered by Dr. Carl Woese from the University of Illinois in 1977. Before that, it was thought that life had two major domains: eukaryotes (e.g., plants, animals and fungi) and bacteria. Although archaea and bacteria are very similar in size and shape, archaea often occur in various shapes, from spheres (0.1 micrometers in diameter) to rods or other random shapes. Despite having no nucleus, archaea's biochemical properties are more closely related to those of eukaryotes. It was thought that some archaeal species had evolved into eukaryotes after diverging from bacteria.
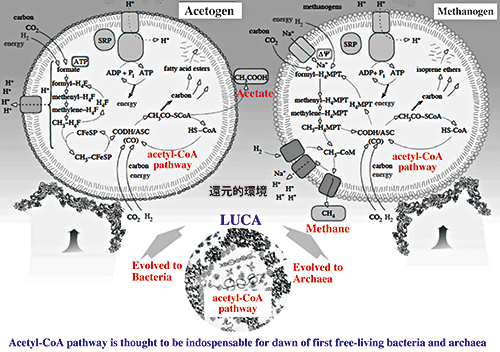
Figure 1. Schematic diagram for the scenario of early life evolution
This figure was slightly modified from the original picture, which appeared in the article of Martin and Russell, Phill. Trans, R. Soc. B (2007) 362, 1887-1926. In the environment of early Earth, acetyl-CoA pathway leading carbon fixation and energy generation is thought to have been indispensable for the emergence of the first free-living bacteria and archaea, which had evolved from LUCA. It is hypothesized that bacteria and archaea were evolved from LUCA by the acquisition of acetogenic and methanogenic energy generation systems, respectively.
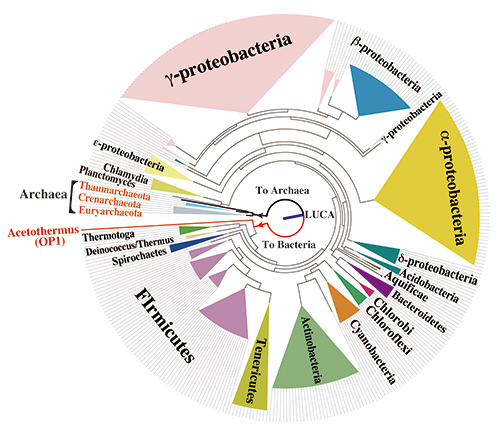
Figure 2. Phylogetetic tree of prokaryotes based on the concatenated amino acid sequence of 4 common proteins
Bar in the center circle shows the LUCA. This phylogenetic tree shows that Ca. 'A. autotrophicum' is a most deeply branching bacterium among the 358 bacterial species whose genome sequences have been determined to date.
In addition, researchers constructed another phylogenetic tree based on the concatenated amino acid sequence of 5 core proteins of the acetyl-CoA pathway. This result also supports that Ca. 'A. autotrophicum' is a most deeply branching bacterium similar to the former phylogenetic tree (Fig. 3B)
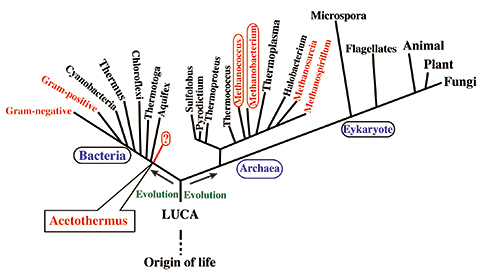
(A) Phylogenetic tree based on the ribosomal RNA genes. Red character shows the organisms possessing acetyl-CoA pathway. Deeply branching acetogenic bacteria possessing the acetyl-CoA pathway has not yet been found in the bacterial lineage in contrast to the methanogenic archaea shown by the red circle.
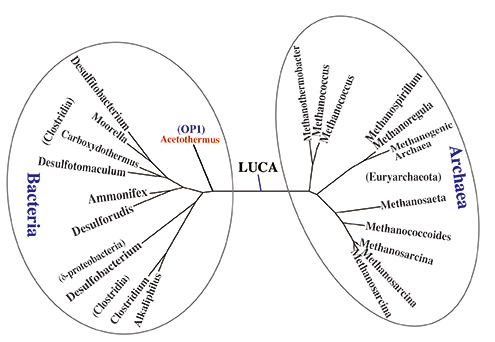
(B) Phylogenetic tree based on the concatenated amino acid sequence of 5 core proteins for the acetyl-CoA pathway. It has been confirmed that Ca. 'A. autotrophicum' diverged first from the common ancestral lineage among the nine examined bacteria.
Figure 3. Phylogenetic tree of life
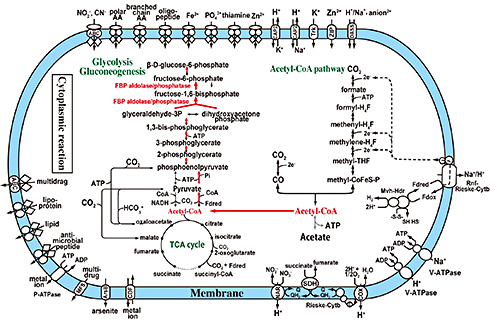
Fig.4. Acetyl-CoA and gluconeogenetic pathways found in Ca. 'A. autotrophicum'.
Red arrows show the direction of gluconeogenesis from acetyl-CoA. This figure suggests that Ca. 'A, autotrophicum' can acquire energy by producing acetate from acetyl-CoA and also synthesize saccharides from acetyl-CoA under anaerobic conditions.
Contacts:
- Japan Agency for Marine-Earth Science and Technology
(For the study) - Hideto Takami, Team Leader
Environmental MetaGenome Research Team,
Extremobiosphere Research Program
Institute of Biogeosciences (BioGeos) - (For publication)
- Hikaru Okutsu, e-mail: press@jamstec.go.jp
Senior Administrative Specialist, Planning Department Press Office